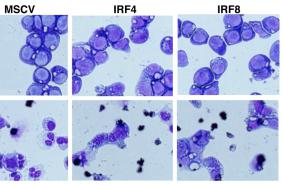
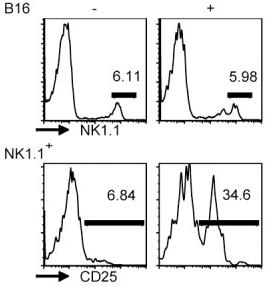
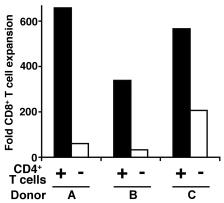
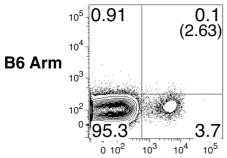
Mouse GM-CSF Recombinant
Categories: HematopoietinsIL-3 familyRecombinant Mouse Cytokines$70.00 – $2,900.00
Description
Accession
P01587
Source
Optimized DNA sequence encodingMouse Granulocyte Macrophage-Colony Stimulating Factor mature chain was expressed in Escherichia Coli.
Molecular weight
Native Mouse Granulocyte Macrophage-Colony Stimulating Factor isgenerated by the proteolytic removal of the signal peptide and propeptide, this molecule has a calculatedmass of kDa. Recombinant mouse GM-CSF is a monomer protein, consisting of amino acidsandmigrates as an approximately14 kDa protein under reducing conditions.
Purity
>95%, as determined by SDS-PAGE and HPLC
Biological Activity
The ED50 as determined by the dose-dependent stimulation of the proliferation of murine FDC-P1 cells is ≤.1ng/ml, corresponding to a specific activity of ≥3 x units/mg.
Protein Sequence
MWLQNLLFLG IVVYSLSAPT RSPITVTRPW KHVEAIKEAL NLLDDMPVTL NEEVEVVSNE FSFKKLTCVQ TRLKIFEQGL RGNFTKLKGA LNMTASYYQT YCPPTPETDC ETQVTTYADF IDSLKTFLTD IPFECKKPGQ K
Endotoxin
Endotoxin content was assayed using a LAL gel clot method. Endotoxin level was found to be less than 0.1 ng/µg(1EU/µg).
Presentation
Recombinant mouse GM-CSF was lyophilized from.2 μm filtered PBS solution, pH7.0.
Reconstitution
A quick spin of the vial followed by reconstitution in distilled water to a concentration not less than 0.1 mg/mL. This solution can then be diluted into other buffers.
Storage
The lyophilized protein is stable for at least years from date of receipt at -20° C. Upon reconstitution, this cytokine can be stored in working aliquots at2° -8° C for one month, or at -20° C for six months, with a carrier protein without detectable loss of activity. Avoid repeated freeze/thaw cycles.
Usage
This cytokine product is for research purposes only.It may not be used for therapeutics or diagnostic purposes.
Molecular function
Molecular function
Methods
In-vitro NK-DC co-culture
- Whole blood samples were obtained from healthy laboratory donors for the isolation of purified and enriched NK and DC cell populations from PBMC using magnetic cell separation.
- The development of an NK-DC co-culture system required two independent steps.
- Firstly, DC were prepared using a pre-optimised magnetic cell sorting kit for the extraction of CD14+ monocytes from PBMC.
- Following isolation, CD14+ monocytes were treated with 100 ng/ml of GM-CSF and 1000 U/ml of IL-4 in RPMI containing 5% autologous serum and maintained in a 5% CO2 cell culture incubator at 37°C for 5 days.
- This altered their phenotype to that of immature dendritic cells (iDC) with a population of >90% CD14−CD1a+ iDC cells, comparable with the published literature +CD16+ NK cell.
- Purities of >98% were attained consistent with published literature 2 cell culture incubator at 37°C.
- Co-culture cell ratios…
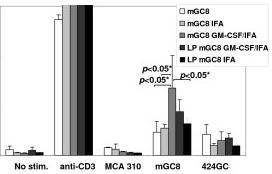
Tumor-specific IFN-γ release and cell-mediated cytotoxicity after vaccination with mGC8 cells and GM-CSF.
- Vaccination with mGC8 with or without LP, GM-CSF, and IFA.
Tumor cell vaccination (prophylactic/therapeutic), LRAST
- To determine the immunogenicity of the tumor cells, 107 tumor cells were irradiated with 10,000 rad and subcutaneously injected into mice.
- Two weeks later, the mice were challenged by subcutaneous injection of 3 × 106 viable tumor cells into the opposite flank.
- Experimental groups generally consisted of 5 mice.
- Tumor development was followed by serial measurements of the tumor diameter and is depicted as tumor size (mm2) = d × D, where d and D were the shortest and the longest tumor diameter, respectively.
- Animals were euthanized when D reached 10 mm.
- Lymphopenia was induced by i.p.
- injection of cyclophosphamide (200 mg/kg, , ).
- This dose was chosen since earlier studies have shown an increased proliferation and long-term survival of antigen-specific T cells at this dose of cyclophosphamide, alone or in combination with fludarabine [7 naïve syngeneic splenocytes followed by s.c. vaccination with irradiated mGC8 cells (107, 10,000 rad) with or without a…
BMDC culture
- Bone marrow was extracted from the femur and tibia passed through a screen and washed in cold media.
- 2×106 bone marrow cells were resuspended in 10 ml of DC media (RPMI, 10% LPS free FCS, β-mercaptoethanol, L-glutamine, 40 ng/ml GM-CSF and cultured in 10 cm petri dishes at 37°C.
- On day 3, 10 ml of DC media was added to the culture.
- On day 6 and 8, 10 ml of culture was removed, resuspended in 10 ml of fresh DC media and added back to the culture.
- On day 9, BMDCs were resuspended in DC media− (ex GM-CSF) at 2×106cell/ml, re-plated in 24 well plates and stimulated with LPS (100 ng/ml), Poly I∶C (100 µg/ml), and/or IFNα (1000 u/ml).
- On day 10, BMDCs were pulsed for 2 hours with gp33 peptide alone or gp33, gp276 and gp61 peptides (10−6 M) respectively, and washed prior to use in
in…
Immunoprotection analysis of Com1- and HspB-pulsed BMDCs
- Mouse bone marrow dendritic cells (BMDCs, CD11c+) were isolated from the bone marrow of BALB/c mice, according to the protocol described previously [2.
- Approximately 1 × 106 cells was added to each well of a six-well plate, and mouse GM-CSF (20 ng/ml) and IL-4 (10 ng/ml) was added to the culture medium every other day.
- After 6 days of culture, BMDCs were stimulated with
C. - burnetii I Ag (10 μg/ml), Com1 (10 μg/ml), HspB (10 μg/ml),
E. - coli LPS (2 μg/ml), or 25 μl elution buffer (mock pulse) for 24 h at 37°C and 5% CO2.
Generation of BMDC and BMMC
- Complete medium'>medium'>RPMI (cmedium'>medium'>RPMI), medium'>medium'>RPMI 1640 medium containing 10% fetal calf serum , was used as culture medium.
- For BMDC induction, 5×106 BM cells were cultured supplemented with 10 ng/ml recombinant murine GM-CSF for five days
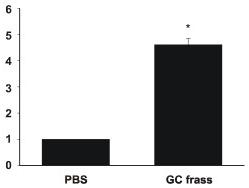
GC frass-induced PAR-2 mRNA and cytokine production from BMDC.
- Bone marrow was isolated from BALB/c mice and cultured in the presence of GM-CSF for 6 days.
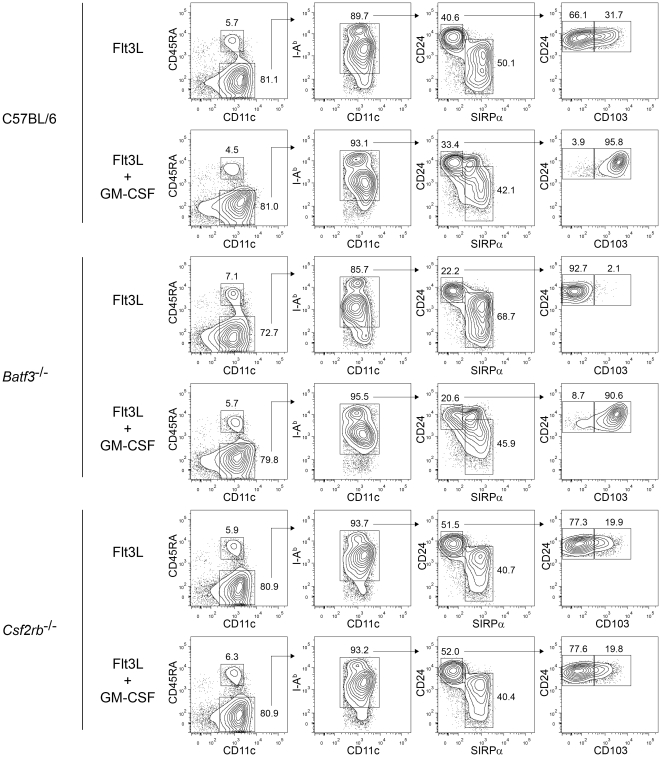
GM-CSF induces CD103 expression on Flt3L-derived CD8α-equivalent DCs.
- FACS analysis of developing DCs in bone marrow cells from C57BL/6, Batf3
Phagocytic cells assays
- The monocyte/macrophage J774.3 cell line used in this study was kindly provided by Dr. María Inés Becker .
- J774.3 cells were routinely grown in high-glucose medium'>DMEM medium (GIBCO), supplemented with 10% Fetal Bovine Serum and 1 mM HEPES (GIBCO) in T75 bottles.
- Cells were incubated at 37°C and 5% CO2 until 95% of confluence.
- Before infection assays, cells were treated with 0.1 mg/ml trypsine for 5 min, recovered in 50 ml polypropylene tubes, and centrifuged at 1,800× g for 5 min at room temperature.
- After three washes with supplemented DMEM medium, cell number and viability was determined in a haemocytometer, using the trypan blue staining (1 mg/ml).
- 5×105 cells/ml were seeded in 24 well-plates and incubated overnight at 37°C and 5% CO2.
- DCs were prepared from bone marrow precursors of C57BL/6 mice.
- Cells were incubated in complete RPMI 1640 medium supplemented with 5% FCS…
Generation of bone marrow-derived DCs
- Mouse BMDCs were generated from bone marrow progenitor cells, according to the modified protocol of[