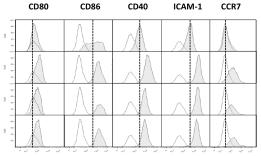
Human beta Nerve Growth Factor Recombinant
Categories: Neurotrophic factorsRecombinant Human Cytokines$80.00 – $1,000.00
Description
Accession
P01138
Source
Optimized DNA sequence encoding Human Nerve Growth Factor mature chain was expressed in CHO cells.
Molecular weight
Native human Nerve Growth Factor is generated by the proteolytic removal of the signal peptide and propeptide the molecule has a calculated molecular mass of approximately 13 kDa. Recombinant human NGF is as a homodimeric protein consisting of two 118 amino acid residue subunits, and migrates as a 13 kDa protein under reducing conditions in SDS-PAGE.
Purity
>95%, as determined by SDS-PAGE and HPLC
Biological Activity
The ED(50) was determined by the determined by its ability to proliferate TF-1 cells and was found to be in the range of.5 ng/ml.
Protein Sequence
MSMLFYTLIT AFLIGIQAEP HSESNVPAGH TIPQAHWTKL QHSLDTALRR ARSAPAAAIA ARVAGQTRNI TVDPRLFKKR RLRSPRVLFS TQPPREAADT QDLDFEVGGA APFNRTHRSK RSSSHPIFHR GEFSVCDSVS VWVGDKTTAT DIKGKEVMVL GEVNINNSVF KQYFFETKCR DPNPVDSGCR GIDSKHWNSY CTTTHTFVKA LTMDGKQAAW RFIRIDTACV CVLSRKAVRR A
Endotoxin
Endotoxin content was assayed using a LAL gel clot method. Endotoxin level was found to be less than 0.1 ng/µg(1EU/µg).
Presentation
NGF-beta was lyophilized from a 0.2 μm filtered solution inmM sodium acetate,mM NaCl, pH.5.
Reconstitution
A quick spin of the vial followed by reconstitution in distilled water to a concentration not less than 0.1 mg/mL. This solution can then be diluted into other buffers
Storage
The lyophilized protein is stable for at least years from date of receipt at -20° C. Upon reconstitution, this cytokine can be stored in working aliquots at2° -8° C for one month, or at -20° C for six months, with a carrier protein without detectable loss of activity. Avoid repeated freeze/thaw cycles.
Usage
This cytokine product is for research purposes only.It may not be used for therapeutics or diagnostic purposes.
Interactor
Interactor
Interactor
Q99523
Interactor
P07174
Interactor
Molecular function
Molecular function
Molecular function
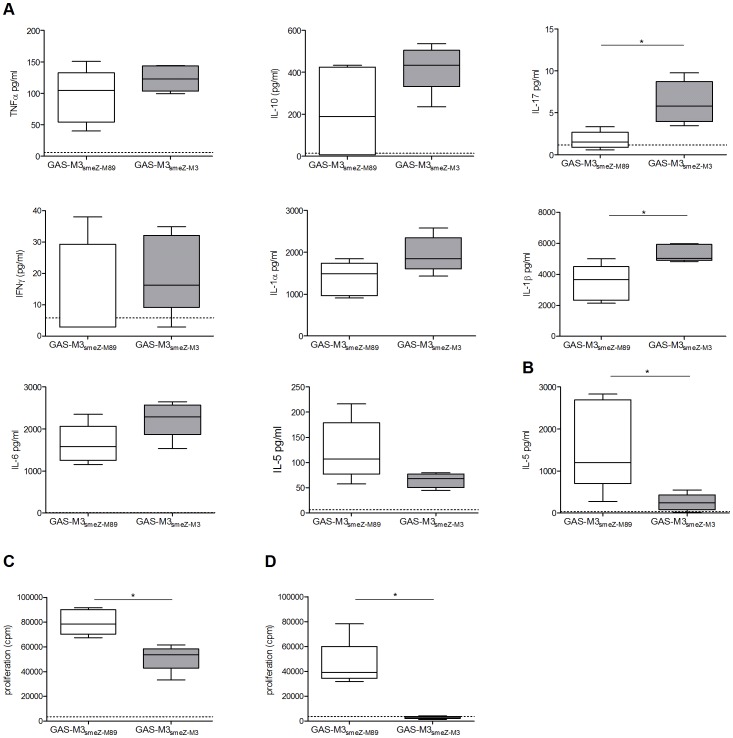
Methods
Migration assay
-
Peripheral blood samples at TP2 in EDTA tubes are diluted with PBS supplemented with 2 mM EDTA and 0.5% BSA (v/v, 1∶1); 30 mL of diluted blood solution is then loaded in a 50-mL LeucoSep tubes with 15 mL of Histopaque 1077 and centrifuged at 850×
g for 25 min. - The buffy coat layers containing white blood cells are collected and washed twice with sterile PBS.
- Cells are then resuspended in EBM-2 supplemented with 0.1% BSA.
- 0.5–1×107 cells/mL of cells are loaded onto the upper part of ThinCert™ tissue culture polystyrene inserts (452.4 mm2 culture surface, 3.0 µm pore size , , ) which are pre-assembled on 6-well plates containing medium as above with or without 100 ng/mL of recombinant human SDF-1α or β-NGF , respectively.
- Cells are then incubated at 37°C in a humidified incubator with 5% CO2 for 18 hrs.
- Cells from both parts of the transwell inserts are dissociated…
- The amount of NGF released from gels with and without the C6S-binding peptide and C6S was quantified.
- Gels (50 µl) were prepared in triplicate in 2 ml siliconized tubes, as previously described, that contained 2 µg/ml NGF (13257019).
- After the gels were cross-linked, 2 ml of 1xPBS was added onto each gel.
- At each time point, 500 µl of buffer was collected from each tube and immediately replaced with 500 µl of buffer using siliconized pipets and tubes.
- NGF release was monitored over 48 h, and samples were stored at -20°C.
- After 2 d, the gels were broken up with a spatula and digested in 10 units/ml collagenase and 0.4 units/ml chondroitinase ABC for 48 h at 37°C with gentle shaking.
- The amount of NGF in the collected samples was quantified with a human β-NGF ELISA development kit (900-K60).
- The absorbance of each well was measured at 405 nm and 650 nm after 10 min…
- The amount of NGF released from gels with and without the C6S-binding peptide and C6S was quantified.
- Gels (50 µl) were prepared in triplicate in 2 ml siliconized tubes, as previously described, that contained 2 µg/ml NGF (13257019).
- After the gels were cross-linked, 2 ml of 1xPBS was added onto each gel.
- At each time point, 500 µl of buffer was collected from each tube and immediately replaced with 500 µl of buffer using siliconized pipets and tubes.
- NGF release was monitored over 48 h, and samples were stored at -20°C.
- After 2 d, the gels were broken up with a spatula and digested in 10 units/ml collagenase and 0.4 units/ml chondroitinase ABC for 48 h at 37°C with gentle shaking.
- The amount of NGF in the collected samples was quantified with a human β-NGF ELISA development kit (900-K60).
- The absorbance of each well was measured at 405 nm and 650 nm after 10 min…